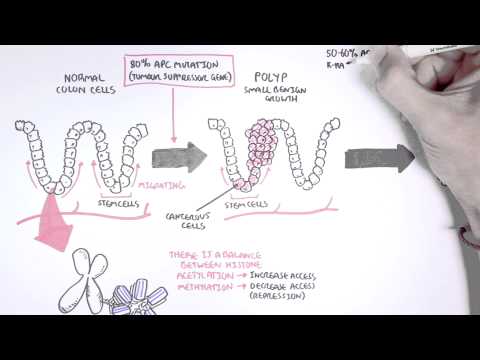
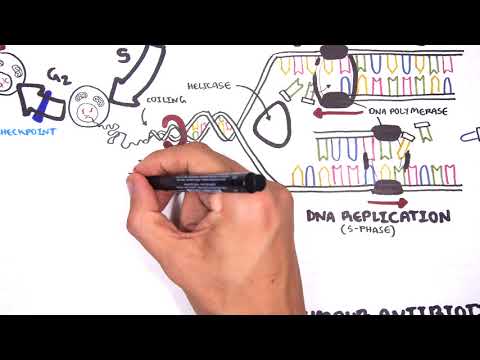
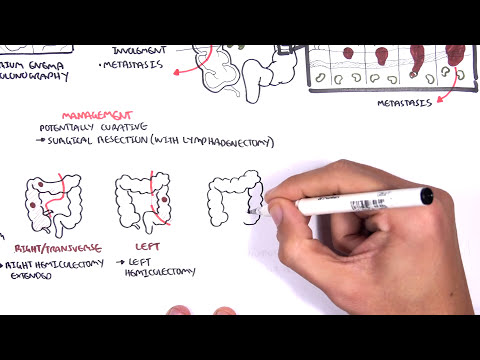
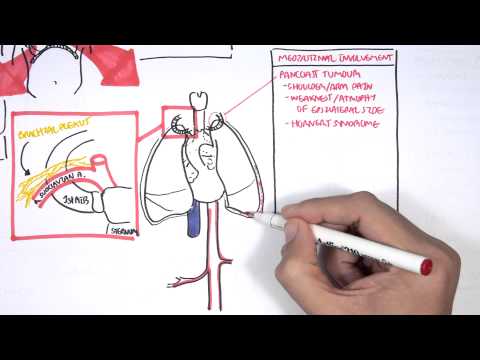
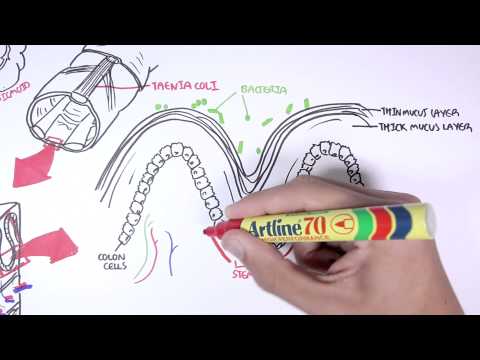
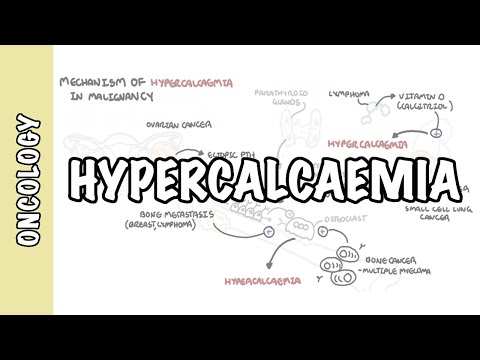
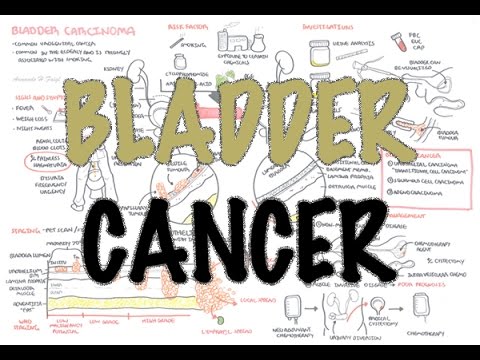
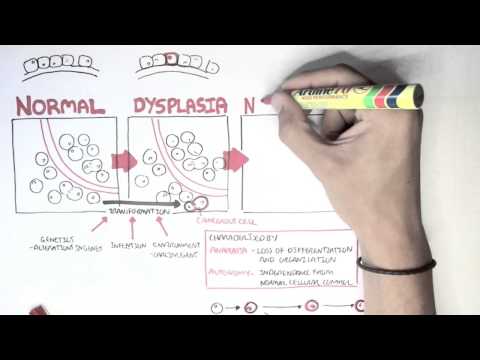
0:00 [MUSIC] 0:05 Despite the huge variation in structure and function, all our cells contain the 0:09 same DNA sequence. 0:11 The reason is that different cells use or express only certain genes. 0:16 DNA can be tagged by tiny chemicals that modify gene expression. 0:21 One of these epigenetic modifications is DNA methylation. 0:26 Promoter DNA methylation is associated with gene silencing, 0:30 and plays an important role in maintaining cell types. 0:36 In cancer, DNA methylation patterns are altered and disrupted. 0:42 DNA methylation is carried out by a group of enzymes known as DNA methyl 0:46 transferases, or DNMT for short. 0:49 We have three major types. 0:52 DNMT1, DNMT3A, and DNMT3B. 0:57 Let's have a closer look at how they work. 1:01 Here we are looking at an early embryonic cell, and we are pulling out its DNA, 1:05 which are wrapped around histone-optimists known as nucleosomes. 1:10 Following fertilization, DNMT3A and DNMT3B are responsible for de novo methyl 1:16 ation, 1:17 allowing embryonic cells to differentiate into a cell type. 1:21 So for example, this embryonic cell has become a distinct cell type, a skin 1:27 cell. 1:28 DNMT1 is responsible for the maintenance of DNA methylation, following 1:34 differentiation, 1:35 and is active during cell division thereafter. 1:42 The methylation patterns of each cell type is different, 1:45 and this reflects the gene expression pattern of the cell. 1:49 So in this case, we have one skin cell becoming many skin cells. 1:55 This cell type has a unique methylation pattern, and therefore expresses 2:02 certain genes. 2:05 Cytosing guining sites, or CPG sites for short, are found all over our DNA. 2:10 For example, here we have a CPG island containing many CPG sites. 2:16 In a normal adult cell, most CPG sites are methylated, except in promoter CPG 2:23 islands. 2:24 These CPG sites are typically un-methylated. 2:28 Promoter regions are regions in the DNA that contain regulatory elements that 2:33 control transcription of genes. 2:40 To understand how CPG sites are methylated, let us zoom into this cytosine 2:45 nucleotide here, 2:46 and look at its chemical structure. 2:49 DNMT3A and DNMT3B are responsible for DNA methylation in early development. 2:57 DNMT obtains the methyl group from a molecule called SAM. 3:01 The methyl group is added here, forming five methyl cytosine. 3:10 It is thought that DNMT flips the cytosine base pair 180 degrees out of the 3:16 strand like so. 3:18 Then, the DNMT enzyme obtains the methyl group from SAM, and transfers it to 3:26 the cytosine. 3:30 Finally, the methylated cytosine is flipped back. 3:40 Human TET, which stands for Human 10/11 Translocation, is another important 3:45 enzyme that has a role 3:46 in regulating DNA methylation patterns. 3:50 TET is responsible for adding a hydroxyl group, initially to five methyl cytos 3:55 ine, forming 3:55 five hydroxyl methyl cytosine. 3:58 The TET enzyme is also able to convert five hydroxyl methyl cytosine back to 4:03 cytosine through several pathways. 4:07 Therefore, the TET enzymes are thought to be responsible for DNA-D methylation. 4:17 In a normal cell, the two opposing processes of methylation and D methylation 4:22 are tightly 4:23 regulated in development. 4:26 However, in cancer, this balance is potentially disrupted, and as a consequence 4:31 , DNA methylation 4:32 patterns change. 4:34 Typically, in cancer cells, we see hypermethylation of promoter CPG islands, 4:40 and this is associated 4:41 with tumor suppressor gene inactivation. 4:45 In contrast to the focal regions of hypermethylation, cancer DNA also undergoes 4:51 widespread high-pole 4:52 methylation across the entire genome. 4:56 This bimodal deregulation of the epigenetic landscape is found in every type of 5:01 human 5:01 tumor. 5:03 Critically, we can now use the alteration in the methylation pattern to help us 5:08 detect 5:09 cancer cells from normal cells.